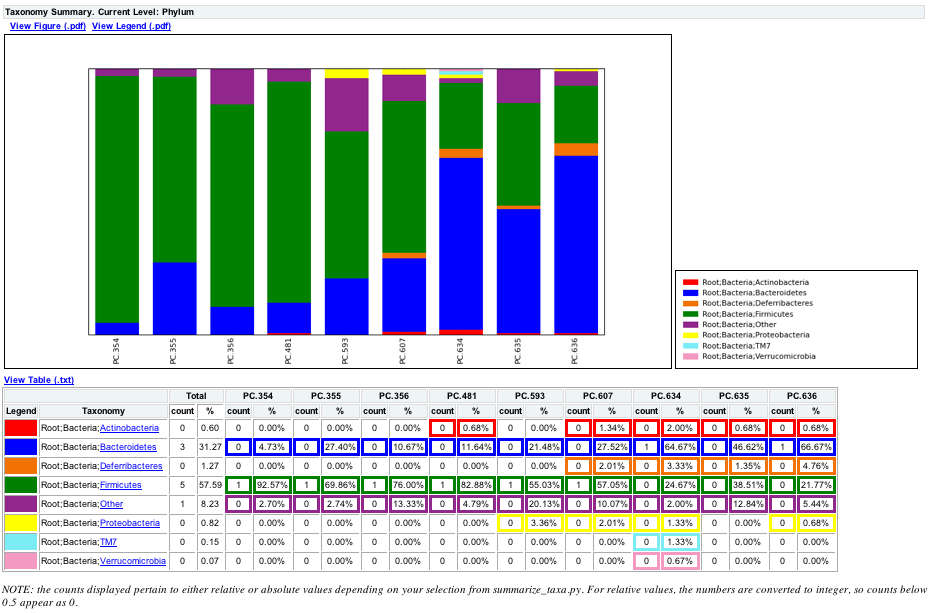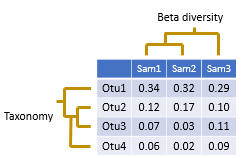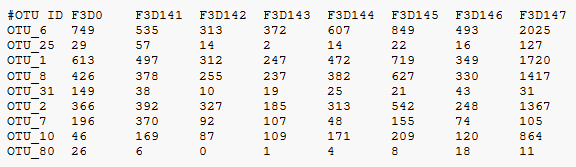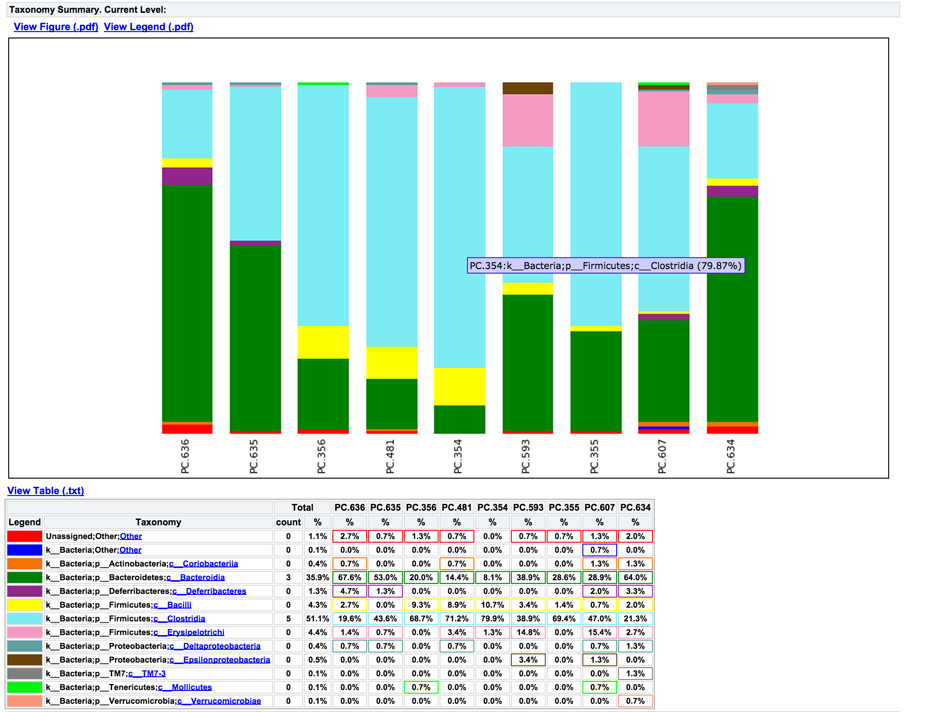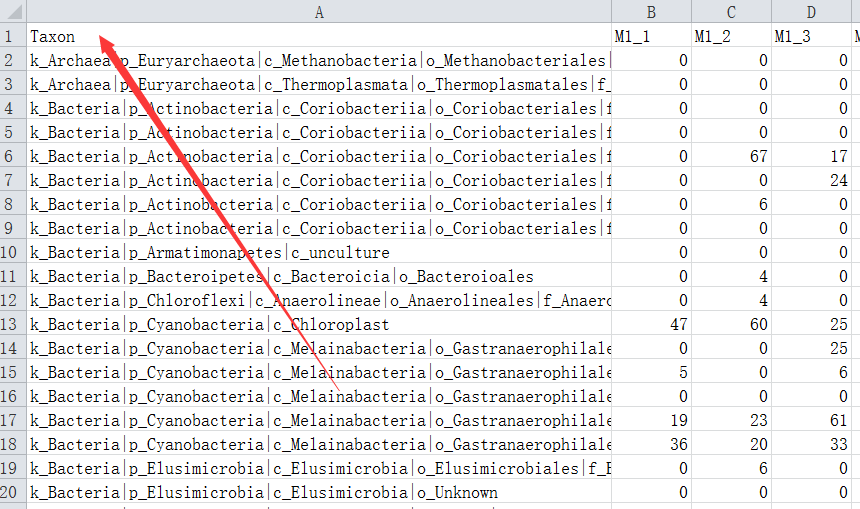
How to add taxonomy to feature-table.qza and get otu id of each taxonomy and number of otus - User Support - QIIME 2 Forum
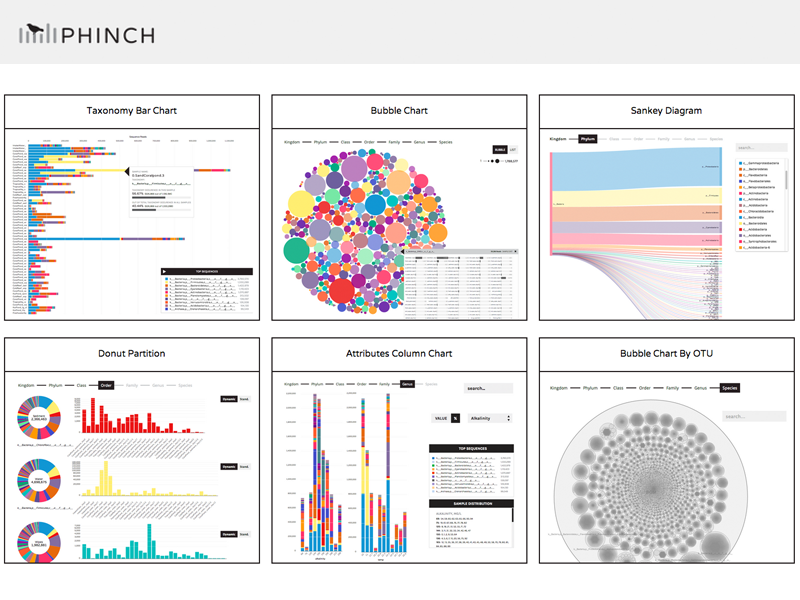
GitHub - PitchInteractiveInc/Phinch: Phinch is an open-source framework for visualizing biological data, funded by a grant from the Alfred P. Sloan foundation. This project represents an interdisciplinary collaboration between Pitch Interactive, a
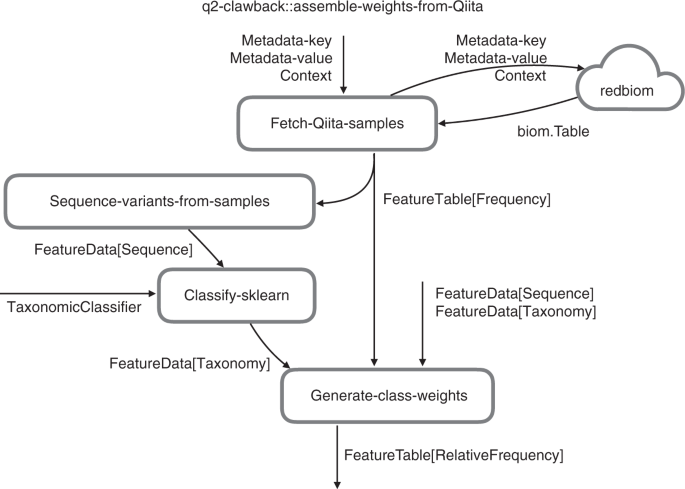
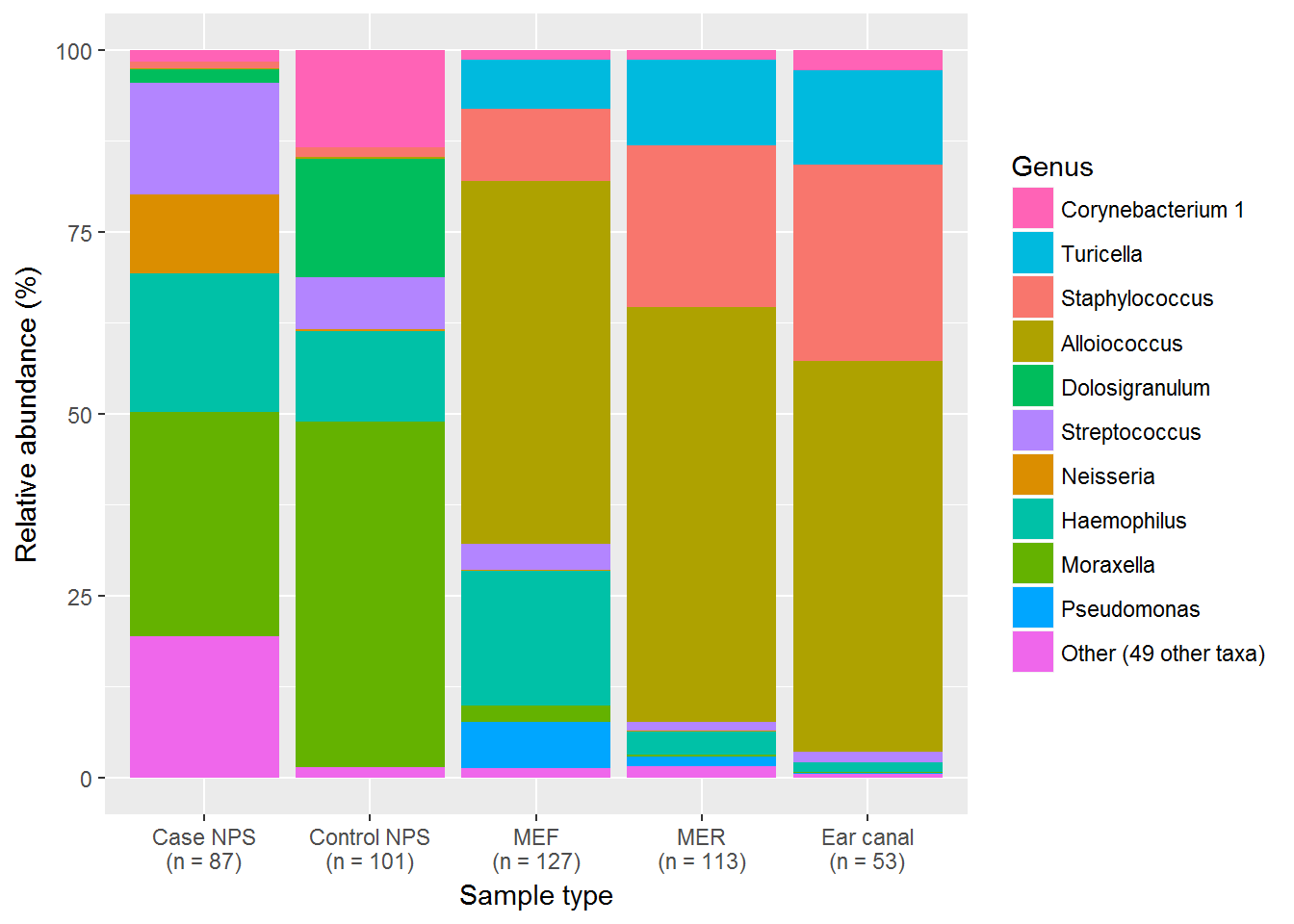
Species abundance information improves sequence taxonomy classification accuracy | Nature Communications

Converting biom files (with taxonomic info) for import in R with Phyloseq - User Support - QIIME 2 Forum