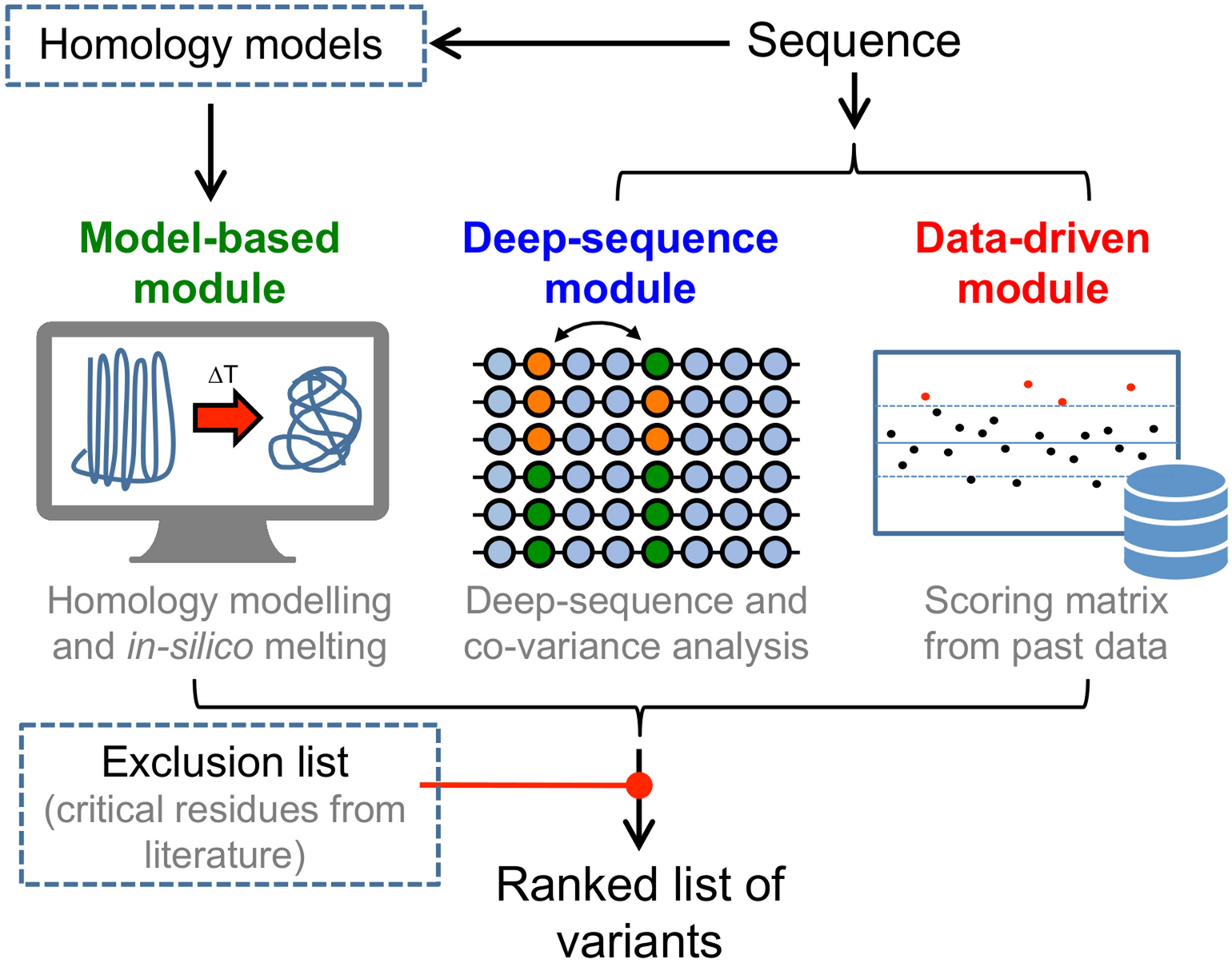
A lipophilicity-based energy function for membrane-protein modelling and design | PLOS Computational Biology
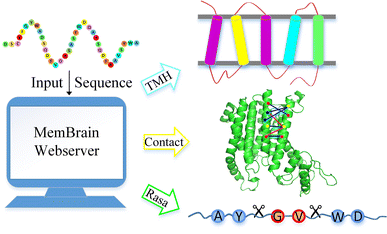
MemBrain: An Easy-to-Use Online Webserver for Transmembrane Protein Structure Prediction | SpringerLink

Methods for Systematic Identification of Membrane Proteins for Specific Capture of Cancer-Derived Extracellular Vesicles - ScienceDirect

Interplay between hydrophobicity and the positive-inside rule in determining membrane-protein topology | PNAS
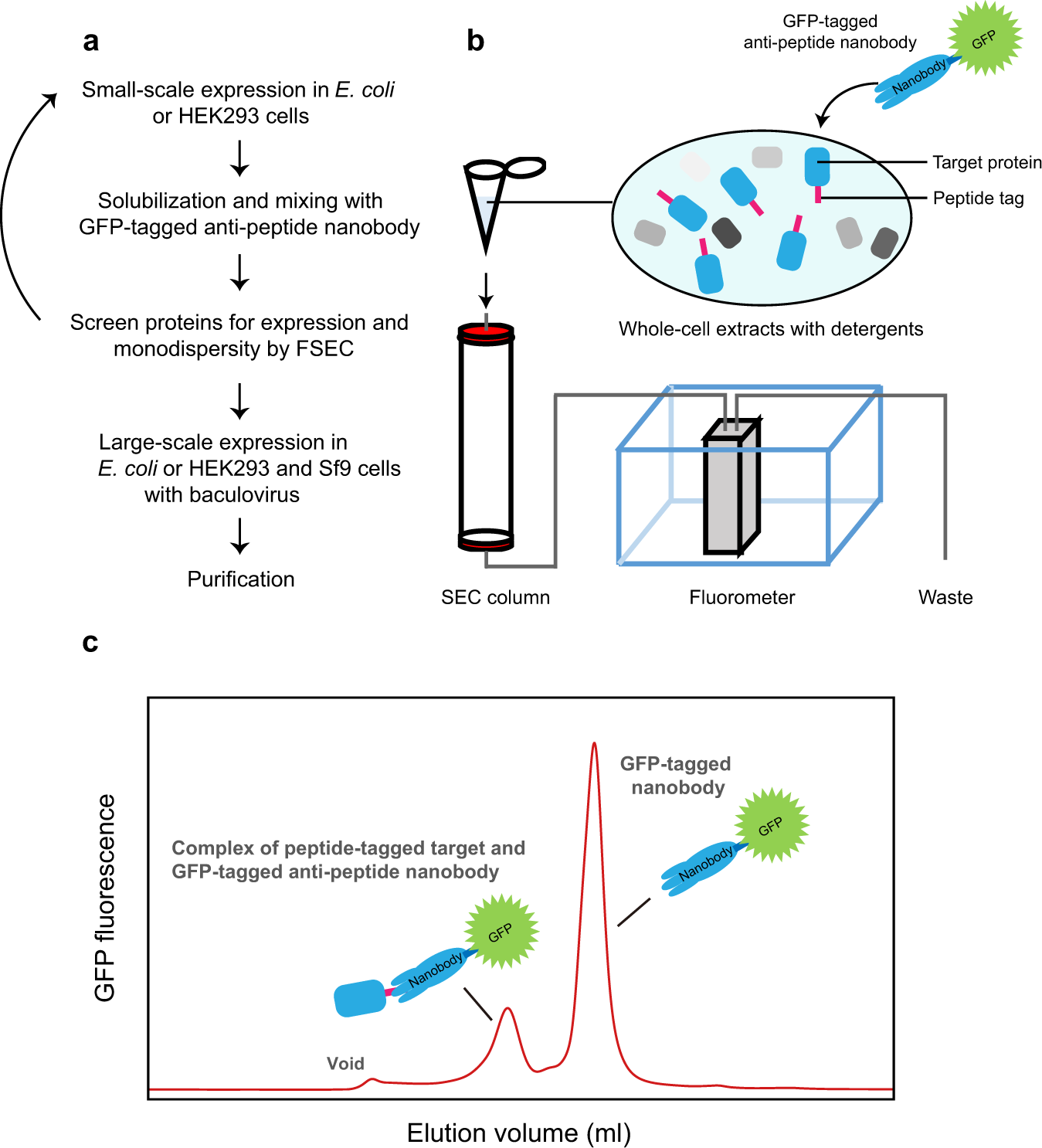
Fluorescence-detection size-exclusion chromatography utilizing nanobody technology for expression screening of membrane proteins | Communications Biology

MemBrain: An Easy-to-Use Online Webserver for Transmembrane Protein Structure Prediction | SpringerLink

Frontiers | A Gene-Based Positive Selection Detection Approach to Identify Vaccine Candidates Using Toxoplasma gondii as a Test Case Protozoan Pathogen

Topology Prediction Improvement of α-helical Transmembrane Proteins Through Helix-tail Modeling and Multiscale Deep Learning Fusion - ScienceDirect

Multiple C2 domains and transmembrane region proteins (MCTPs) tether membranes at plasmodesmata | EMBO reports
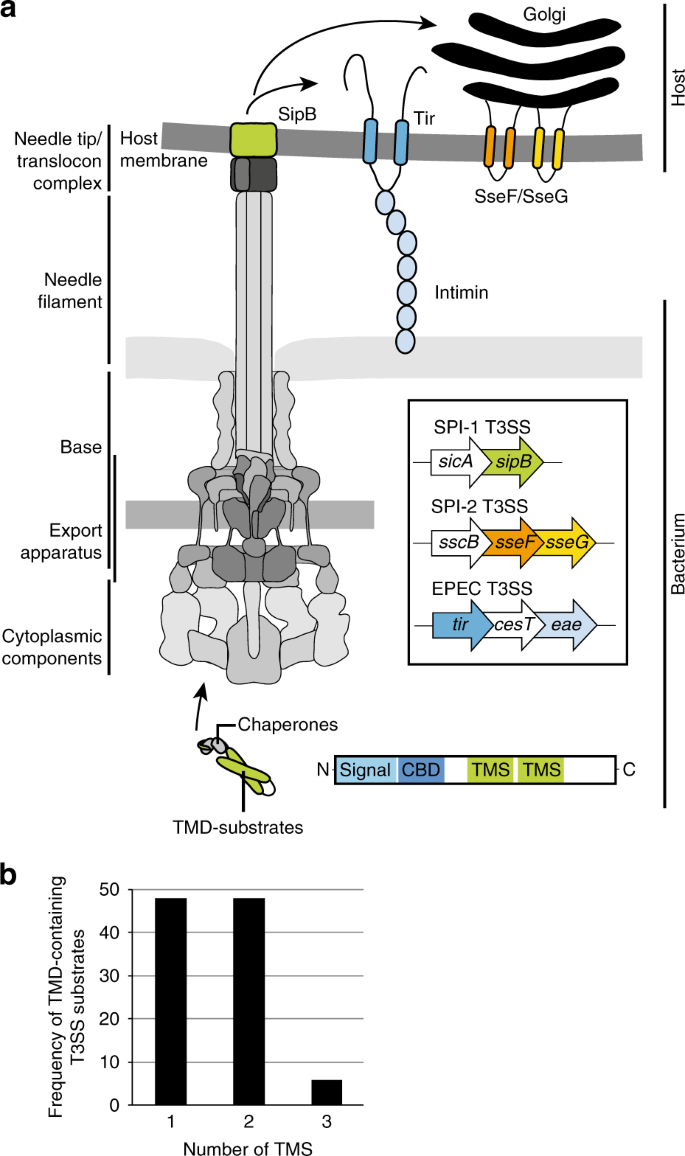
Revealing the mechanisms of membrane protein export by virulence-associated bacterial secretion systems | Nature Communications

Interplay between hydrophobicity and the positive-inside rule in determining membrane-protein topology | PNAS
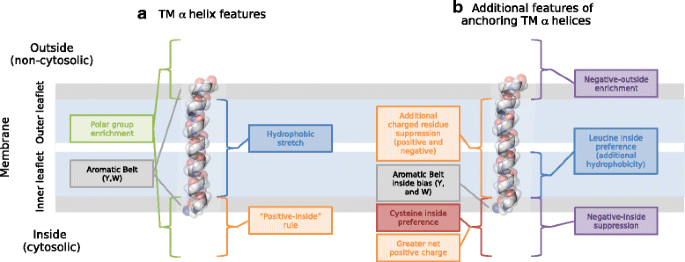
Charged residues next to transmembrane regions revisited: “Positive-inside rule” is complemented by the “negative inside depletion/outside enrichment rule” | BMC Biology | Full Text

Interplay between hydrophobicity and the positive-inside rule in determining membrane-protein topology | PNAS
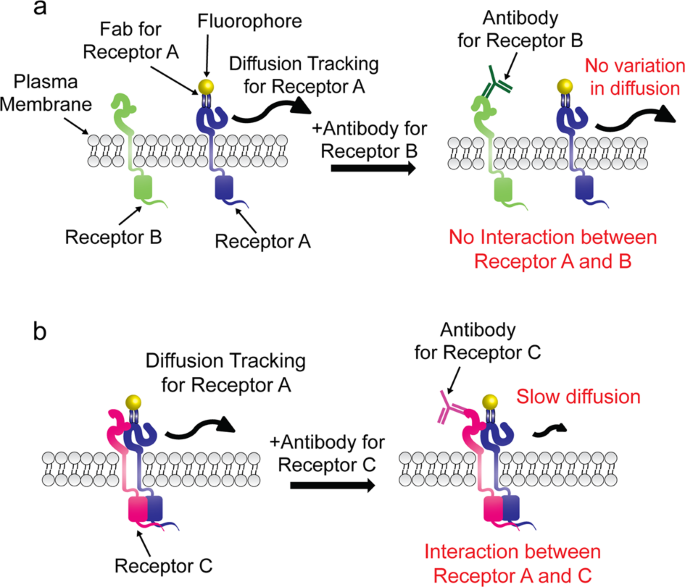
Analysis of transient membrane protein interactions by single-molecule diffusional mobility shift assay | Experimental & Molecular Medicine
Membrane protein contact and structure prediction using co-evolution in conjunction with machine learning | PLOS ONE

Getting to know each other: PPIMem, a novel approach for predicting transmembrane protein-protein complexes - ScienceDirect